¶ Introduction to the bioinfomatics services
The CENTURI Multi-Engineering Platform provide an expertise in Bioinformatics.
Our engineers help scientists to analyse their data, witch are mainly produced by the Next-Generation Sequencing technology. In this context, they develop new analysis pipelines and assist in the design, deployment and user training of data visualization tools. They also promote interactions between the bioinformatics community in Marseille.
¶ Workflow for omics data analysis
Our workflow are available on the Github of the Multi-Engineering Platform.
¶ Microbiome Analysis Workflow: Metabarcoding analysis
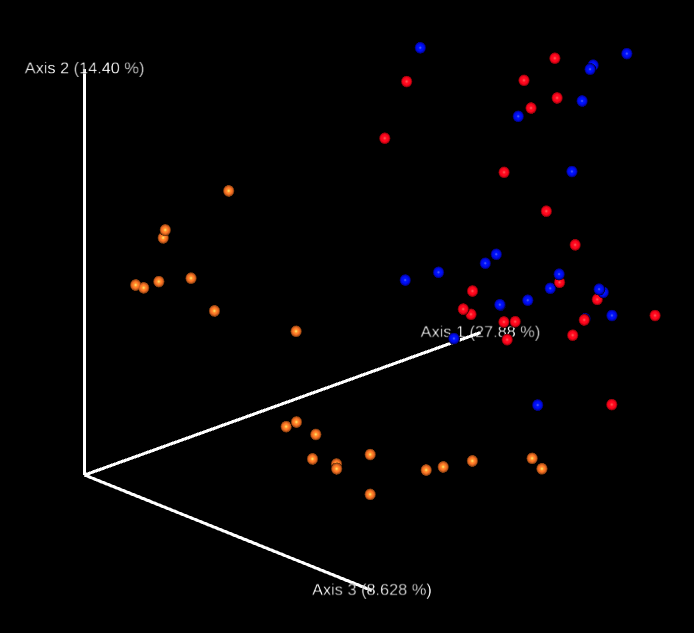
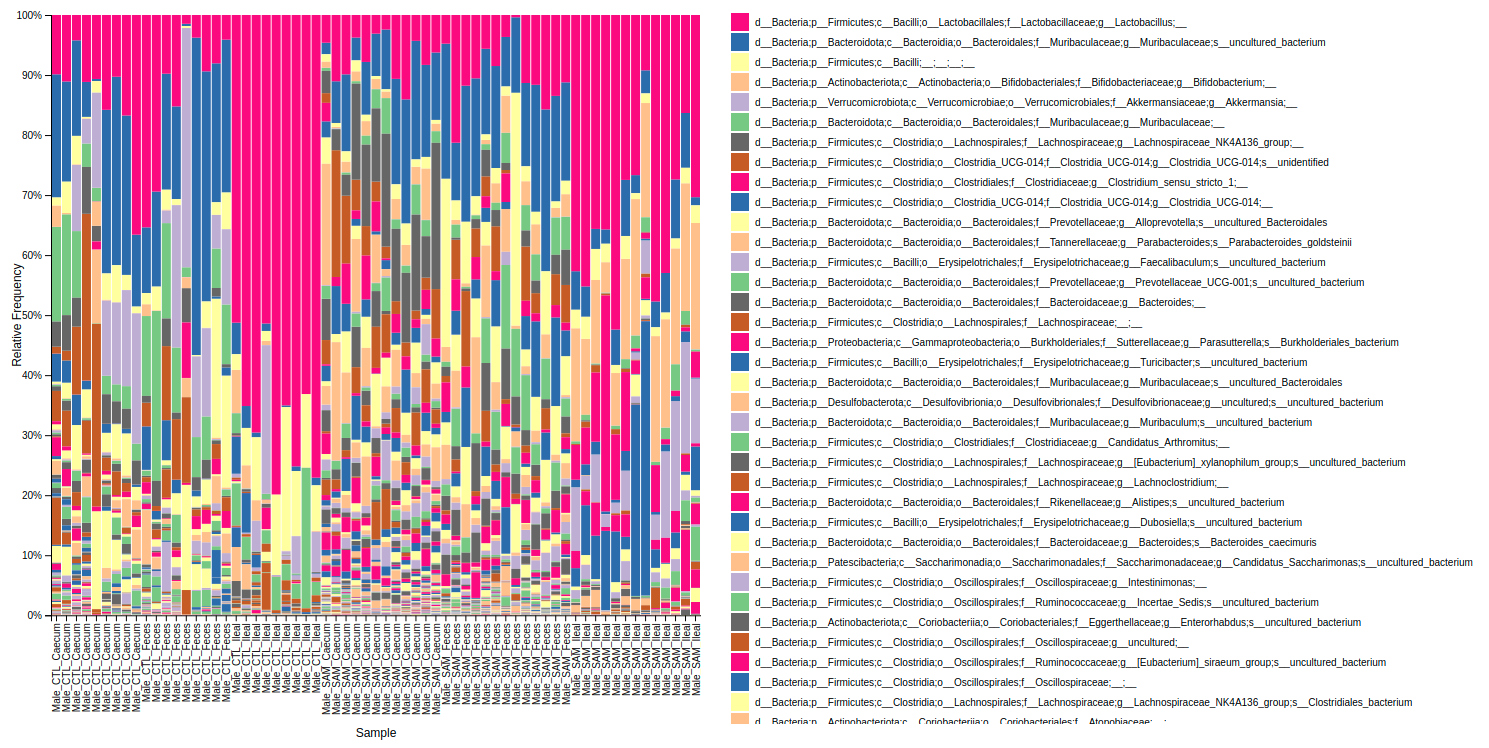
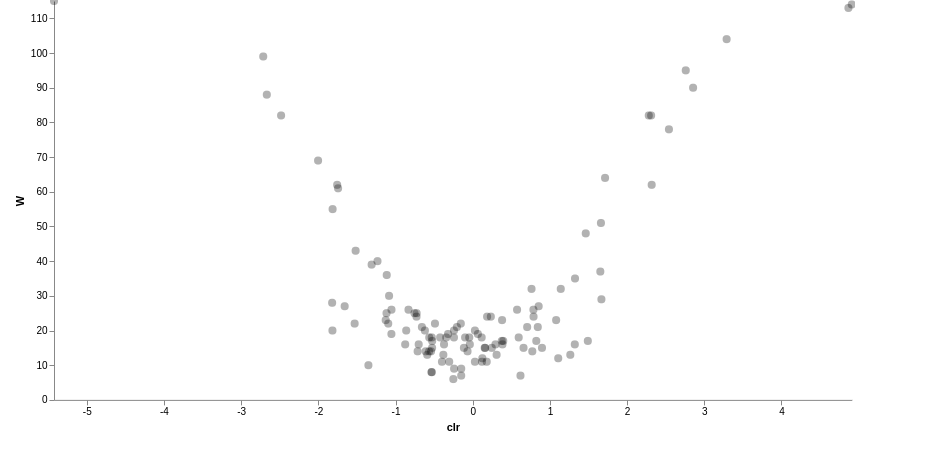
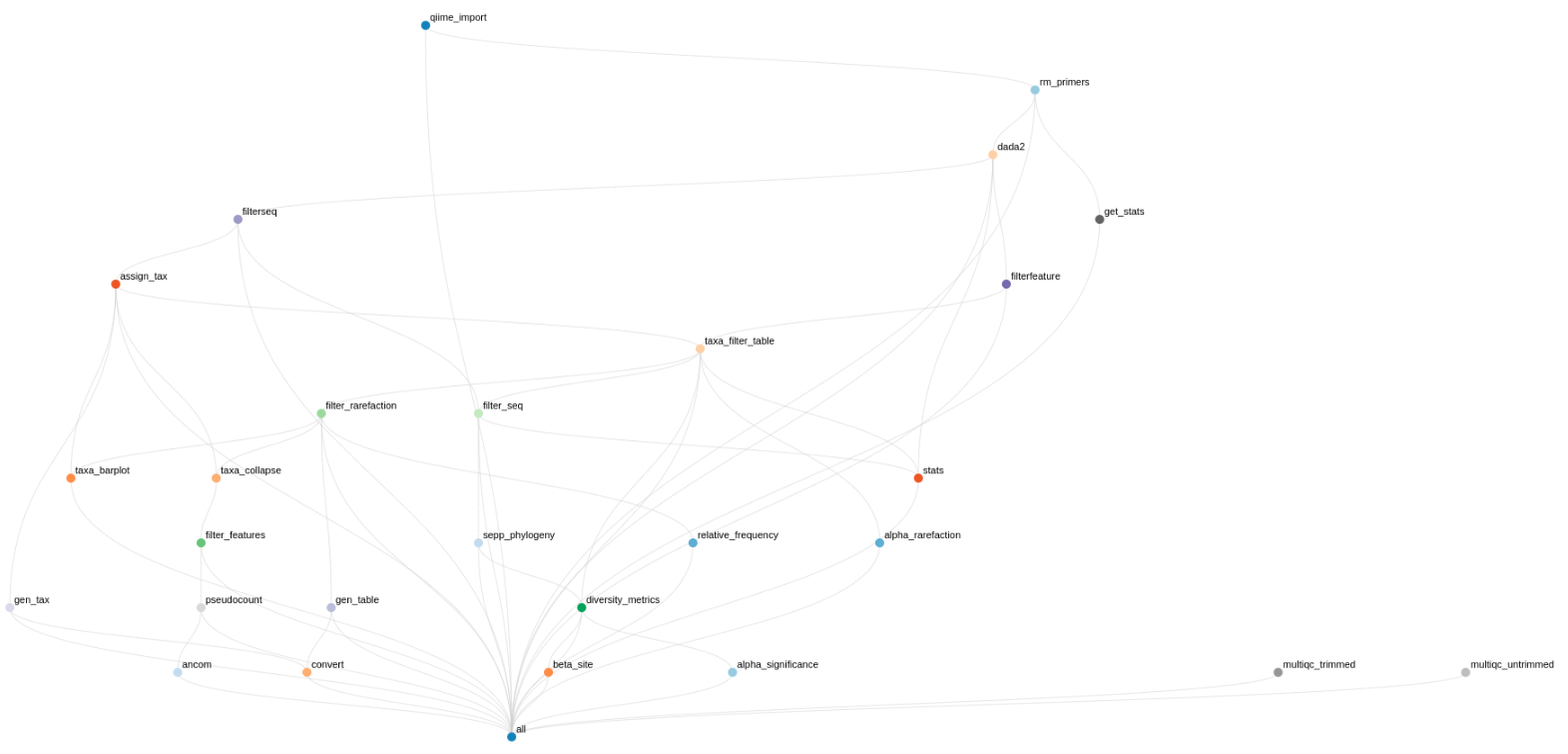
This workflow performs a metabarcoding analysis from fastq files to the production of a report with result of the marker-gene analysis. The analysis are mainly been achieved using plugins from QIIME 2, Bolyen et al., 2019.
You need to install Singularity on your computer. This workflow also work in a slurm environment.
Each snakemake rules call a specific conda environment. In this way you can easily change/add tools for each step if necessary.
Code and supplemental documentations are on Github.
¶ Single-Cell RNA-seq Analysis Workflow
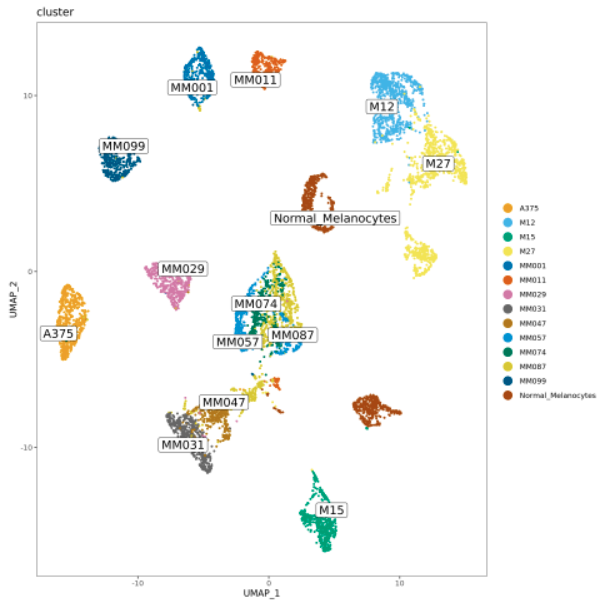
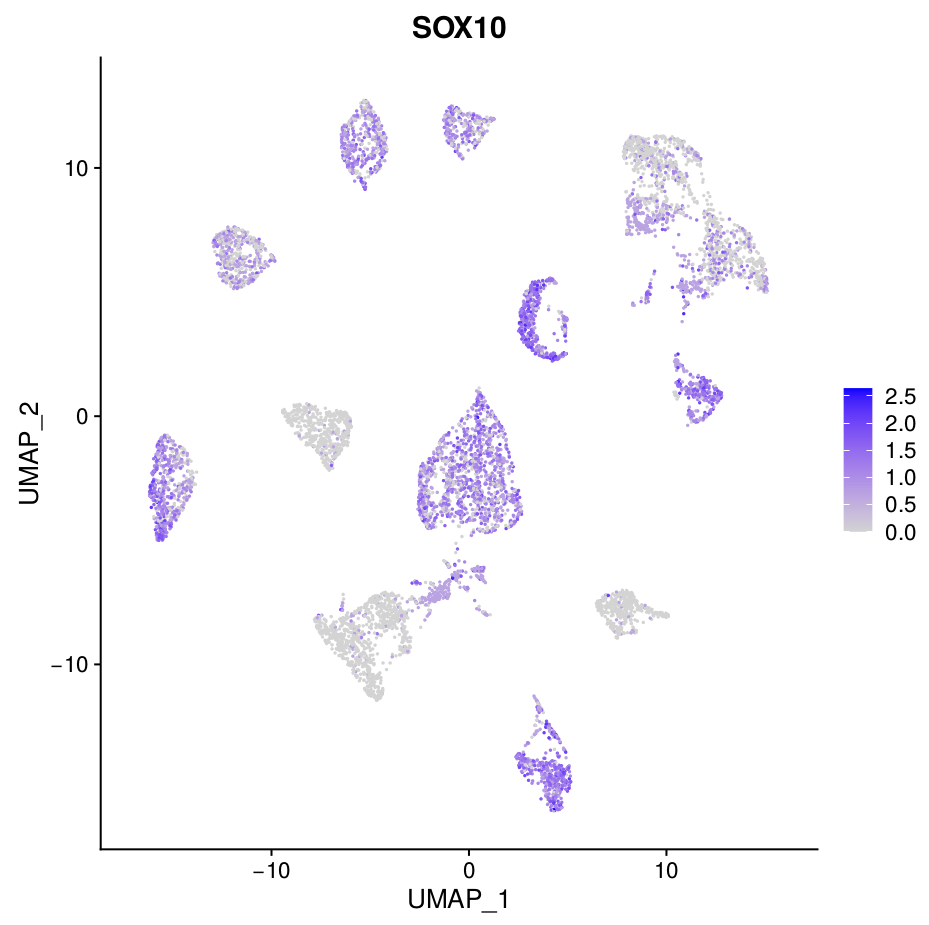
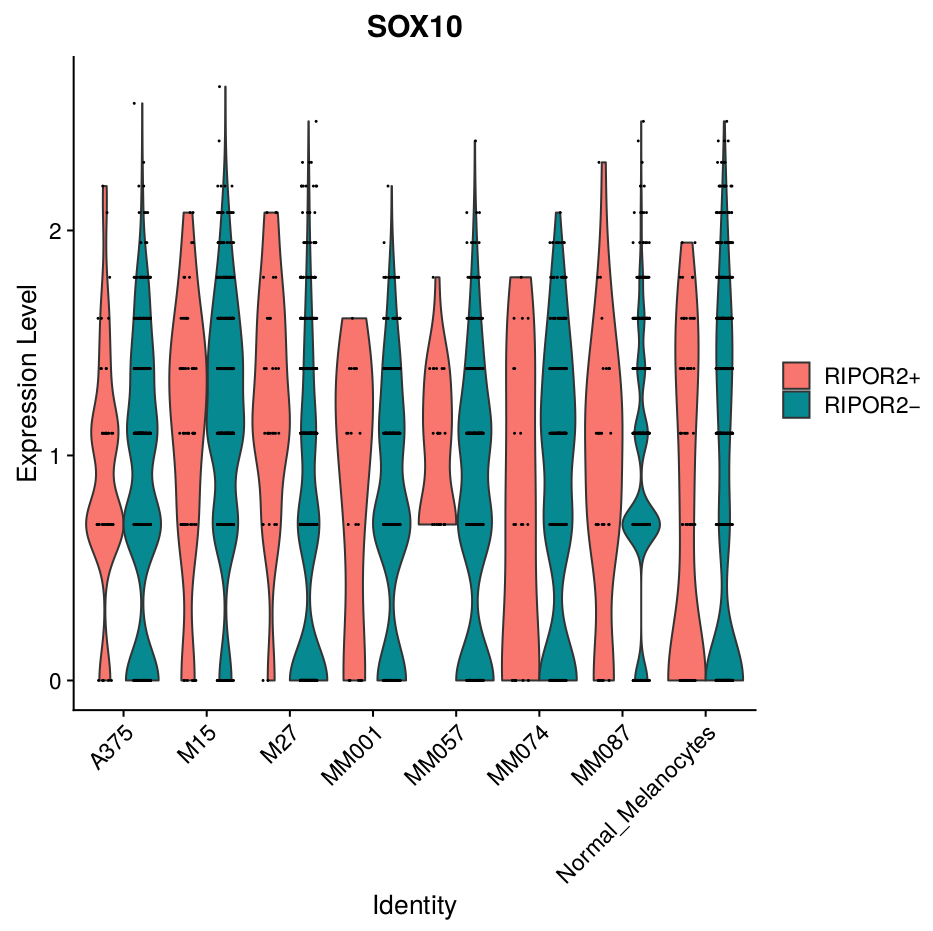
This workflow performs a Snakemake pipeline to process 10x single-cell RNAseq data from fastq files to the analysis of the differential expression of marker-genes. Correction for technical differences between datasets can be included (i.e. batch effect correction) with the integration method during the sctransform process in Seurat to perform comparative scRNA-seq analysis across experimental conditions.
You need to install Singularity on your computer. This workflow also work in a slurm environment.
Steps for the analysis:
- cellranger.smk: Build the reference, count and aggregate with cellranger v6.0.0. You can also do the demultiplexing of your data with cellranger v7.2.0.
- seurat.smk: Run seurat v4.0.3 for the clustering.
- differential_exp.smk: Differential expression based on the non-parametric Wilcoxon rank sum test.
- diffexp_subset.smk: Differential expression analyses inside cluster after a subset on the marker gene expression.
¶ Differential Expression Workflow: RNA-seq analysis
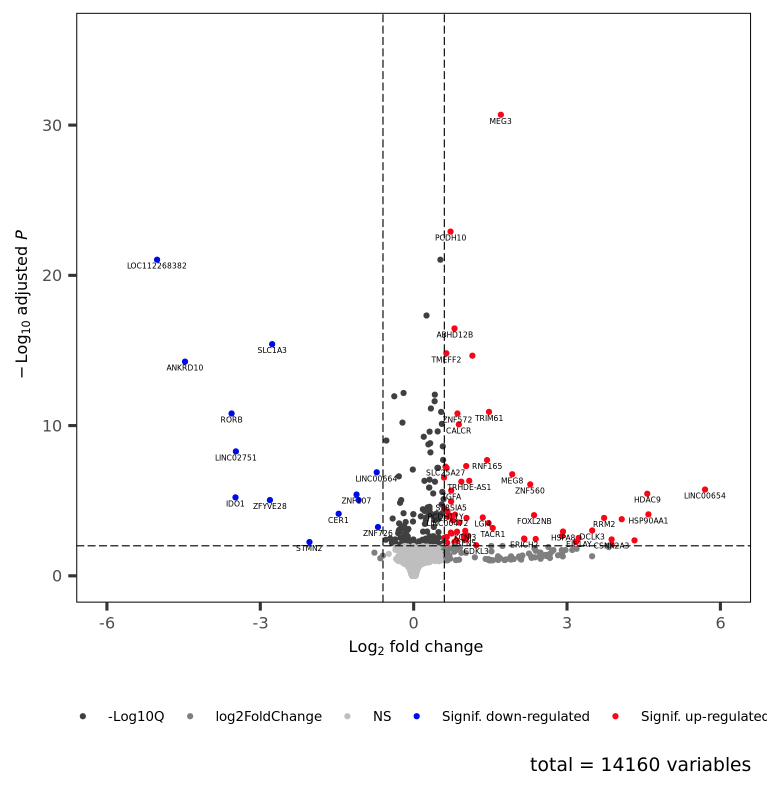
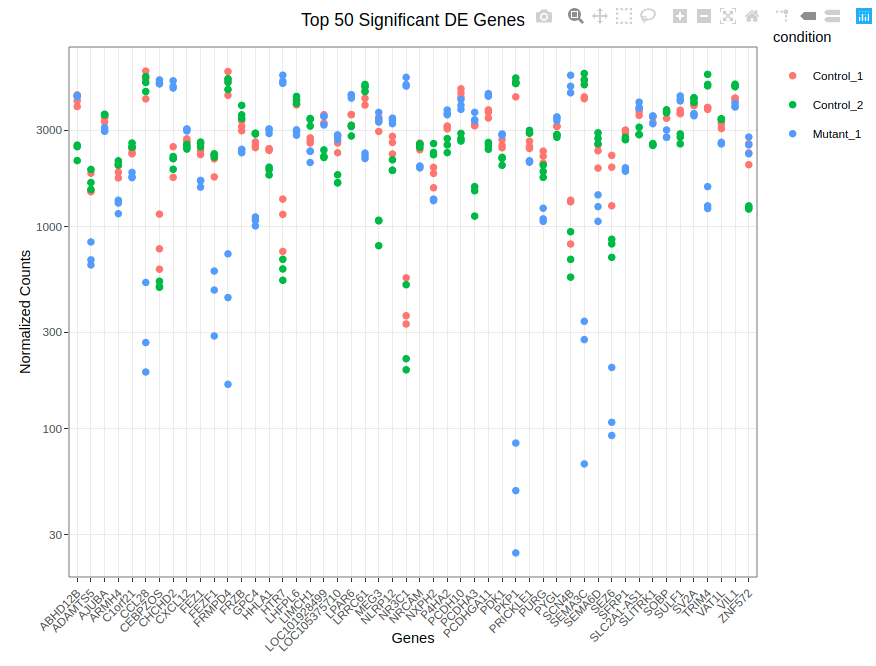
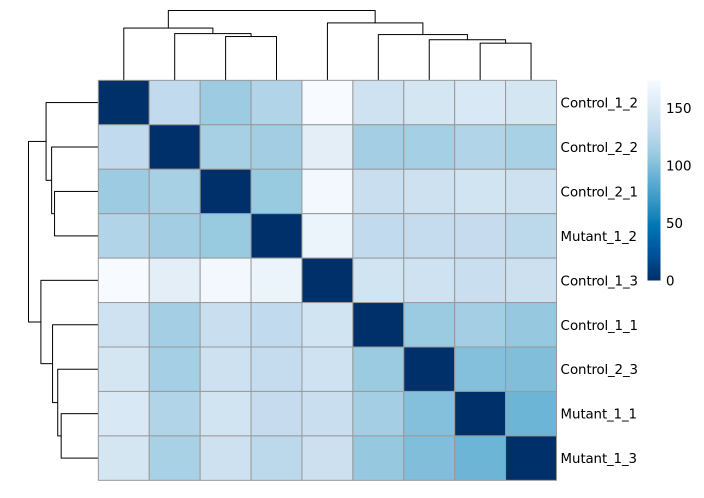
This workflow performs an RNA-seq analysis from the sequencing output data to the differential expression analyses.
It run into a docker container (see Dockerfile) including a general conda environment (see envfair.yaml).
Each snakemake rules call a specific conda environment. In this way you can easily change/add tools for each step if necessary.
3 steps for the analysis:
- clean.smk: The quality of the raw reads are assessed using FastQC v0.11.9 toolkit. Adapters and low quality reads are trimmed using Trimmomatic v0.39.
- count.smk: HiSat2 v2.2.1 is used for mapping the raw reads to the reference genome. The expression for each gene is evaluated using featureCounts from the Subread v2.0.1 package.
- differential_exp.smk: The low expressed genes are removed from further analysis. The raw counts are normalized and used for differential expression testing using DESeq2 v1.28.0.
Code and supplemental documentations are on Github.
¶ Data visualization tools
¶ ShIVA (Shiny Interface for Visualization and Analysis of single-cell datasets)
ShIVA is an interactive software which would allow biologists to be autonomous in the analysis of their single-cell sequencing data, via a graphical interface. This software allows the analysis of data from different single-cell sequencing technologies (scRNA-seq, CITE-seq), used in different disciplines: immunology, developmental biology, neuroscience, and others.
The ShIVA project was initiated and led by the CIML bioinformatics platform.
This tools will be online very soon !
¶ Bioinformatics HUB in Marseille
¶ Bioinformatics Slack channel
This channel encourage interaction between our bioinformatics community on the Luminy campus/Aix-Marseille.
To Join this channel, you can fill this form.
¶ Bioinformatics expertise
Updated 25 June 2021
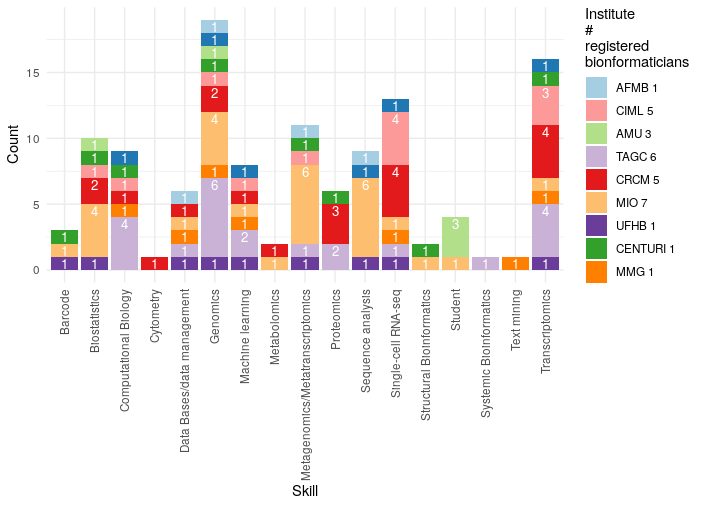
This graph was generated from the answers to a form sent to the bioinformaticians present in some institutes of Marseille. (32 Bioinformaticians fill up the form).This is NOT representative of all the skills present in Marseille!
